
Top-down proteomics as a high-sensitivity tool to assess donor quality in kidney transplantation
Dinesh Jaishankar1, Aniel Sanchez2, Pei Su2, Jeffrey Huang2, Michael Caldwell2, Indira Pla Parada2, Michael Hollas2, Nathaniel Henning2, Michelle A Callegari1, Meredith E Taylor1, Amna Daud1, David Pinelli1, Vinayak Rohan1, Eleonora Forte2, Neil Kelleher2, Satish N Nadig1.
1Surgery, Northwestern University, Chicago, IL, United States; 2Chemistry, Northwestern University, Evanston, IL, United States
Introduction: Despite significant advances in pediatric kidney transplantation (pKTx), donor kidney management remains a major limitation to allograft longevity. Currently, donor kidneys are assessed, in part, by gross inspection, perfusion pump parameters, and histology. While effective, these are not inclusive nor correlative to the discard rates. Therefore, novel, reliable, and sensitive methods of assessing donor kidney quality are crucial to improving donor kidney utilization in pKTx. Top-down proteomics (TDP), a robust methodology using mass spectrometry and bioinformatics, has gained popularity for biomarker development. This methodology allows for identifying proteoforms, i.e., protein isoforms, providing a detailed landscape of the complexities associated with donor kidney management. Here, we investigated whether TDP can be a suitable tool for detecting proteoform landscapes in KTx patients.
Methods: Systemic changes in proteoforms were evaluated by TDP in the blood (PBMCs) collected pre- and one-week post-KTx from adult recipients who received kidneys from living donors (LD), donors after brain death (DBD), and donors after circulatory death (DCD). Local changes in proteoforms were evaluated using spatial proteoform profiling in LD, DBD, and DCD kidney biopsies collected at the backbench prior to KTx. TDP and in-house bioinformatics pipelines were used to acquire and analyze proteoform changes among the donor groups.
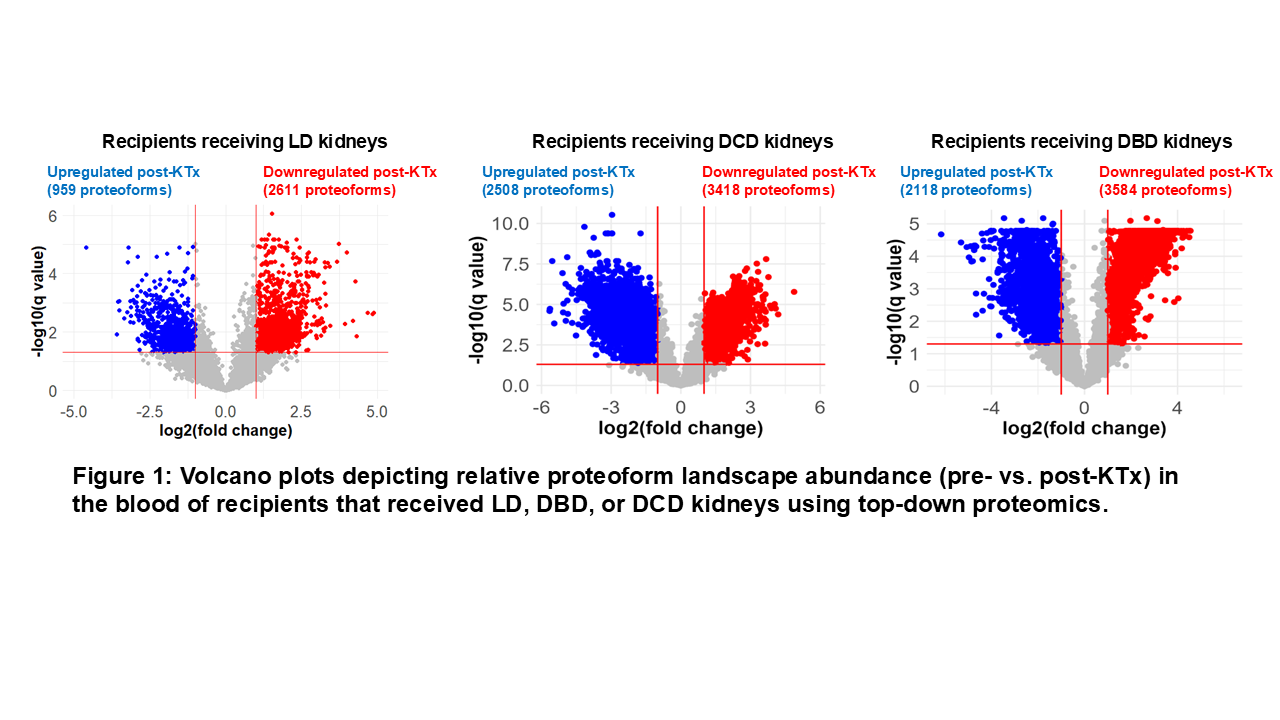
Results: In the blood, the number of proteoforms that changed significantly after KTx was higher in recipients who received DBD/DCD kidneys than those who received LD kidneys, suggesting KTx-associated proteome alternations (Figure 1). The analysis of unique blood proteoforms that were differentially expressed revealed that in recipients receiving DBD/DCD kidneys, proteoforms were more closely associated with an adaptive immune response (antigen presentation) and activation than those receiving LD kidneys, possibly suggesting increased immunological activation in these recipients due to DBD/DCD kidneys. The spatial proteoform profiling of donor kidney biopsies revealed higher abundances of proteoforms corresponding to primary metabolic enzymes observed in LD kidneys, indicating greater metabolic activity. Among the ~100 proteoforms quantified from all samples, proteoforms contributing to cellular structures show a higher abundance in DBD kidneys, possibly correlating with lower mechanical tissue stability in DBD kidneys.
Conclusions: In this pilot study, we successfully demonstrated using TDP to identify and stratify unique proteoform signatures in recipient blood and donor kidney biopsies. Future TDP studies on larger cohorts will further validate and characterize proteoform changes. Additionally, correlating the blood vs biopsy proteoform landscapes will enable biomarker discovery for future point-of-care assays to assess and stratify organ preservation quality in pKTx.
[1] Donor organ quality
[2] Top-down proteomics
[3] Proteoforms