Agnieszka Swiatecka-Urban, United States
Professor of Pedfiatrics, Director of Nephrology
Pediatric Nephrology
University of Virginia
Risk stratification of chronic kidney allograft failure in donor-recipient (D-R) pairs
Wei-Min Chen1, Mateusz Miszczuk2, Alexander Koeppel3, Kenneth L Brayman5, Konstantine Khutsishvili4, Iuliia Vitko2, Shawn Pelletier5, Phillip Ruiz4, Alden Doule4, Agnieszka Swiatecka-Urban1.
1Genome Sciences, University of Virginia, Chaelottesville, VA, United States; 2Pediatrics, University of Virginia, Charlottesville, VA, United States; 3Signature Science LLC, Charlottesville, VA, United States; 4Medicine, University of Virginia, Charlottesville, VA, United States; 5Surgery, University of Virginia, Charlottesville, VA, United States
Study's purpose: Only 1 in 5 chronic kidney graft failures can be attributed to human leukocyte antigen (HLA) mismatches. Unmeasured HLA variants, non-HLA mismatches, and gene polymorphisms affecting drug metabolism influence long-term graft survival. Susceptibility variants are not yet ready for incorporation into clinical practice because their interactions and cumulative risk remain unknown. This study aims to stratify the risk of chronic graft failure based on predisposing genetic modifiers in the D-R pairs.
Methods: We genotyped 114 D-R pairs (IRB-HSR210437) using Global Discovery Array with Enhanced Pharmacogenomic Content (GDA-PGx; Illumina) and generated genome-wide (GWAS) data. We performed a GWAS scan for graft failure using the Cox proportional hazards model and for post-transplant eGFR using the linear regression model. Next, using our in-house toolset SECRET (Screen and Evaluate Catalogued Risk Scores to Enhance Trait prediction), we leveraged the Polygenic Score Catalog to identify polygenic score (PGS) models that predict graft failure and post-transplant eGFR levels. We calculated ~5,000 PGSs for each study subject and then evaluated the power of each PGS to predict graft failure with covariates being adjusted.
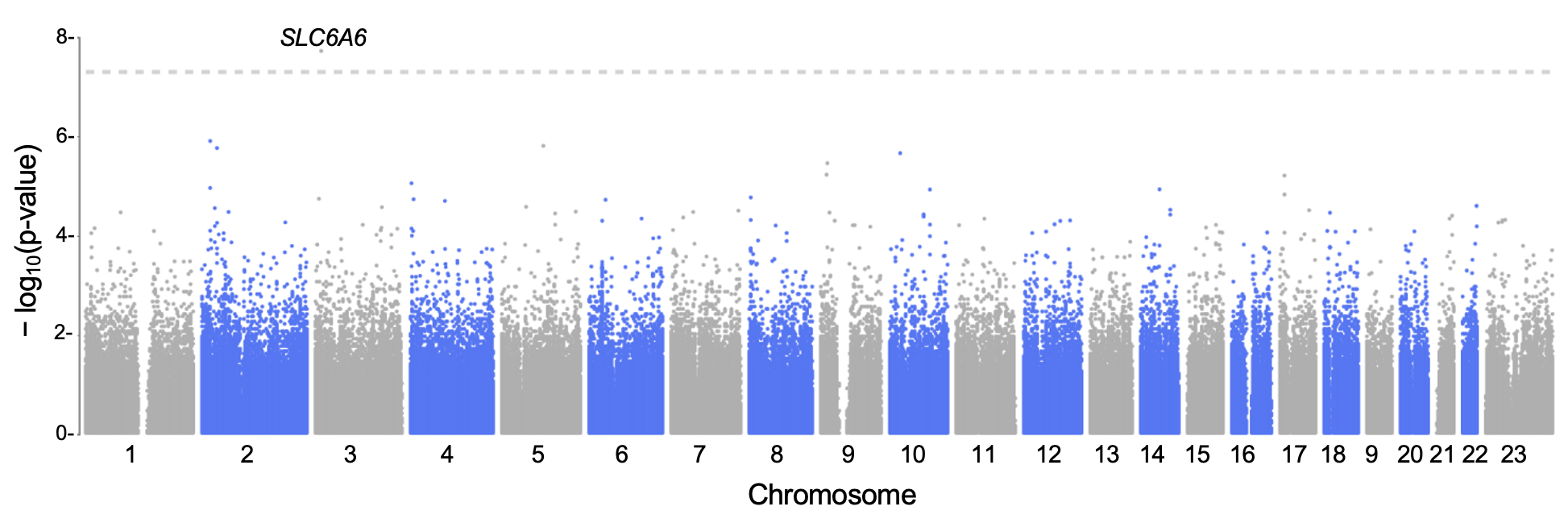
Results: GWAS scan for the graft failure trait identified variant rs62231826 (P= 1.3×10-8), at which the minor allele has a hazard ratio (HR) of 9.3 (Figure 1). This minor allele was also negatively associated with post-transplant eGFR (P=0.002 and P=0.02 for the 1-year and 3-year eGFR). This variant was in the gene SLC6A6, which encodes the sodium-chloride-dependent taurine transporters (TAUT) and has been found in a previous study to predict graft function loss at the level of gene expression, protein abundance, and transporter function. SECRET identified PGS for systolic blood pressure in the donors as the most predictive for graft failure in recipients older than 60 (HR=1.4, P=0.004). The scan identified PGS for HbA1c in the recipient as the most predictive for eGFR decline 5 years after transplant (16% explained by this PGS).
Conclusions: This proof-of-concept pilot study establishes a framework for risk stratification. In the future, fully powered studies may improve graft allocation and survival and provide faster access for those needing kidney transplantation.
National Institutes of Health (NIH) grants R01HL144539 and P50DK096373-11.
References:
[1] Graft failure, polygenic risk score, donor-recipient mismatches, glomerular filtration
Lectures by Agnieszka Swiatecka-Urban
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Fri-19 13:35 - 15:05 |
Improving survival following pediatric kidney transplant | Risk stratification of chronic kidney allograft failure in donor-recipient (D-R) pairs | MOA 6 |
